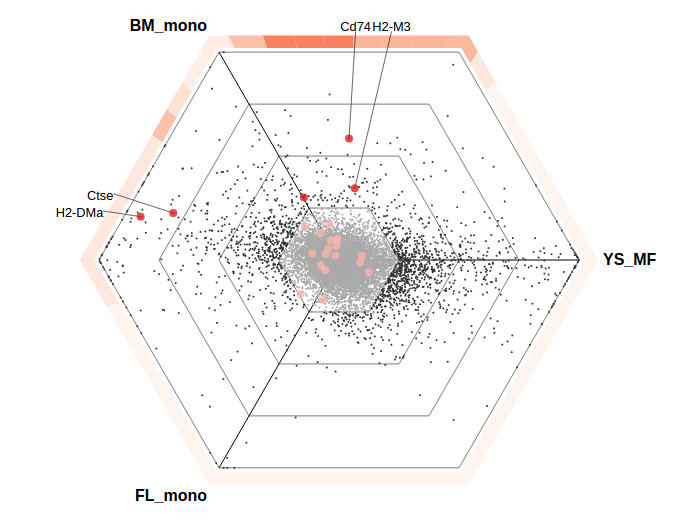
Triwise is an R package to visualize and analyze gene expression data among three biological conditions. Triwise allows the generation of publication-ready plots to give a general overview of the expression differences between the three conditions and certain genes of interest. Triwise can also be used to detect functional gene sets coherently upregulated in one or two of the biological conditions. One application of this method can be found in this study (van de Laar et al. 2016), which is further worked out in the vignette.
The three biological conditions can be related in different ways:
If you have any questions feel free to contact me at wsaelens@ugent.be
van de Laar, Lianne, Wouter Saelens, Sofie De Prijck, Liesbet Martens, Charlotte L. Scott, Gert Van Isterdael, Eik Hoffmann, et al. 2016. “Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages.” Immunity 44 (4): 755–68. doi:10.1016/j.immuni.2016.02.017.